An international study between the Imprinting and Cancer Group within the Cancer Epigenetics and Biology Program (PEBC) and the Institute of Human Genetics, at Christian-Albrechts University, Kiel, Germany, describe a methylation signature that is a promising marker for classifying myeloid-derived leukemias.
Gene Wilms tumor 1 (WT1) is over-expressed in numerous cancers with respect to normal cells, and has either a tumor suppressor or oncogenic role depending on cellular context.
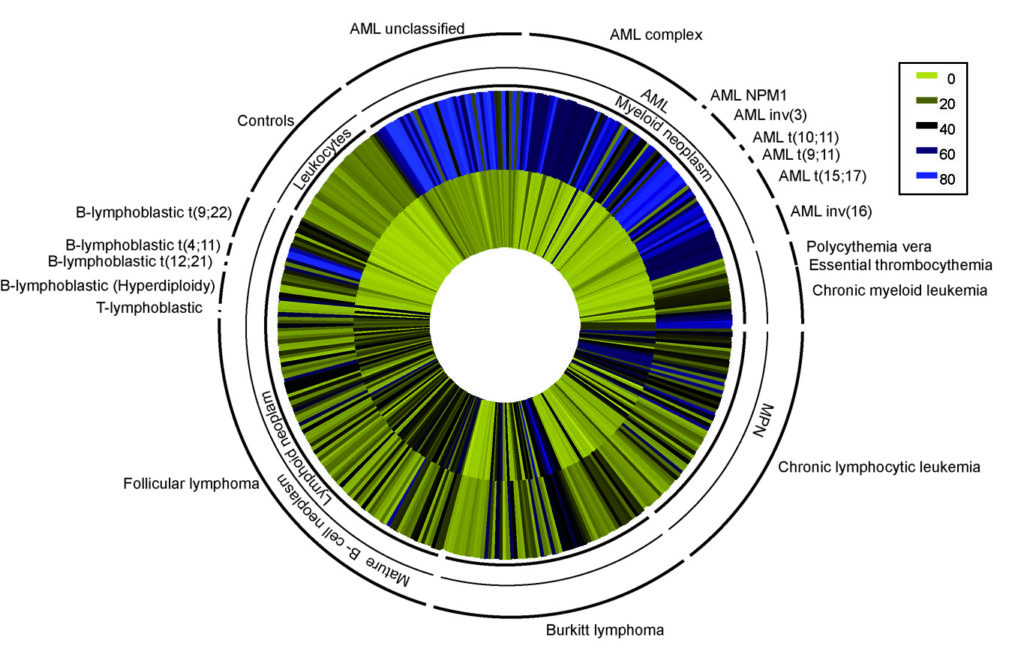
The study published in the Journal Hematology and Oncology (Guiliaumet-Adkins et al., 2014) revealed that despite high expression of WT1 in AML-derived cell lines, there is an epigenetic switch from a permissive to repressive chromatin within the first intron. Subsequent methylation analysis in 356 primary leukemia and lymphoma samples revealed that the epigenetic signature identified in cell lines is specific to myeloid-lineage malignancies, irrespective of underlying mutational status or translocation.
Dave Monk explained that “we show that the robust AWT1 epigenetic signature is an excellent marker for discriminating AML from non-diseased peripheral blood and that this hypermethylation signature can accurately track disease progression, differentiating relapse patients from those achieving complete remission after allogeneic hematopoietic stem cell transplantation (SCT)”.
Hypermethylation of the alternative AWT1 promoter in hematological malignancies is a highly specific marker for acute myeloid leukemias despite high expression levels. Guillaumet-Adkins A, Richter J, Odero MD, Sandoval J, Agirre X, Catala A, Esteller M, Prósper F, Calasanz MJ, Buño I, Kwon M, Court F, Siebert R, Monk D. J Hematol Oncol. 2014 Jan 9;7(1):4.